Check out the new pharmacokinetics data for monoclonal antibodies!
The CAVD and HVTN have seen an increased emphasis on passive immunization studies where participants receive a monoclonal antibody (mAb) as part of a prophylactic or treatment regimen. The PK data from these studies is important for evaluating and further characterizing those mAbs known to be broadly neutralizing (bNAbs), determining the appropriate dose levels required to maintain optimal concentration levels of mAbs in the blood, and identifying potential combinations of bNAbs to use in future studies.
PK mAb data is now available in the DataSpace in a standard and integrated format that facilitates comparisons and meta-analyses across studies.Data from 4 of the 6 completed CAVD studies that collected PK data have been loaded into the DataSpace, and there's more coming!
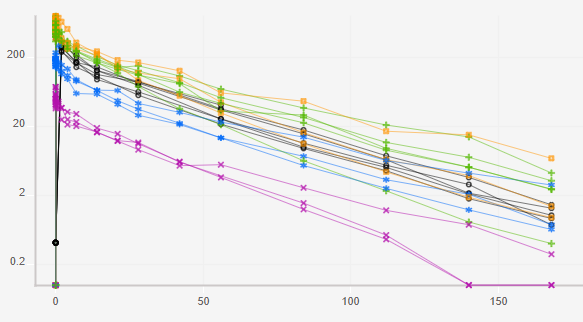
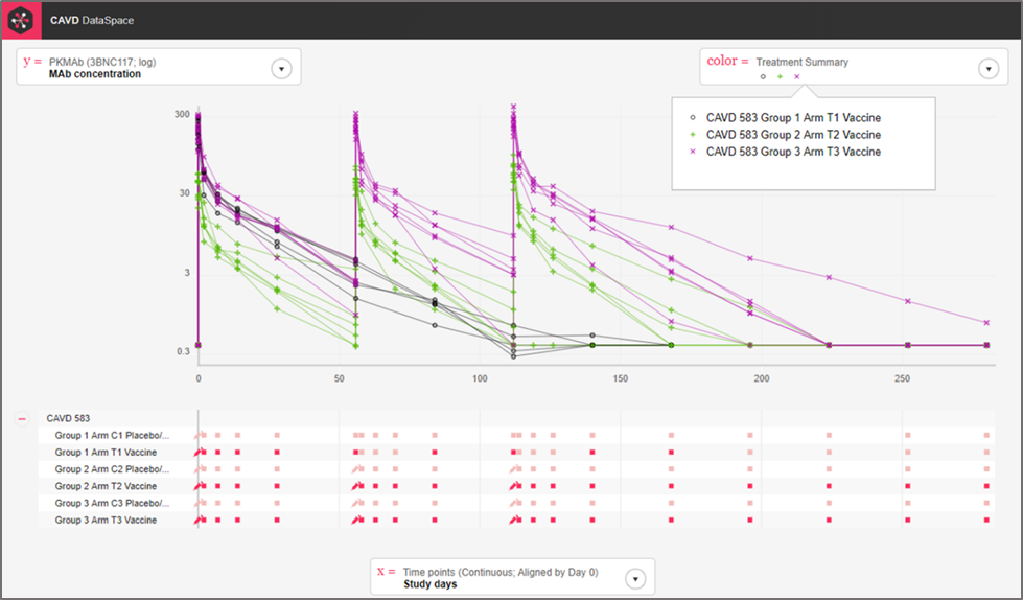
The plot below shows the concentration of 3BNC117 over time for each of the treatment groups in the clinical trial, CAVD 583/Nussenzweig (Protocol YCO-899).
Give it a try!
Step 1: Go to Plot data and select PK MAb as the data source
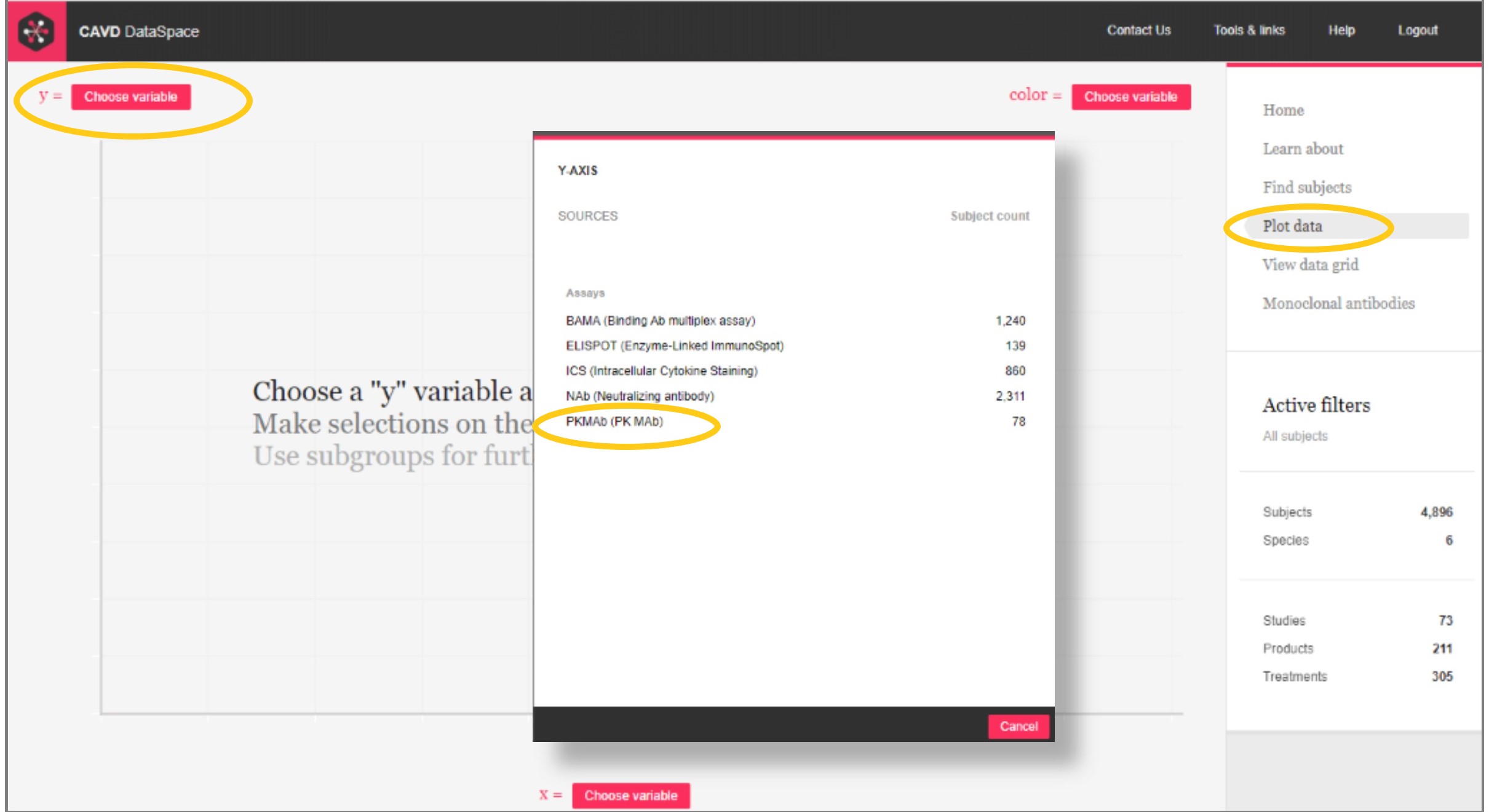
Step 2: Select a mAb to plot and set the y-axis
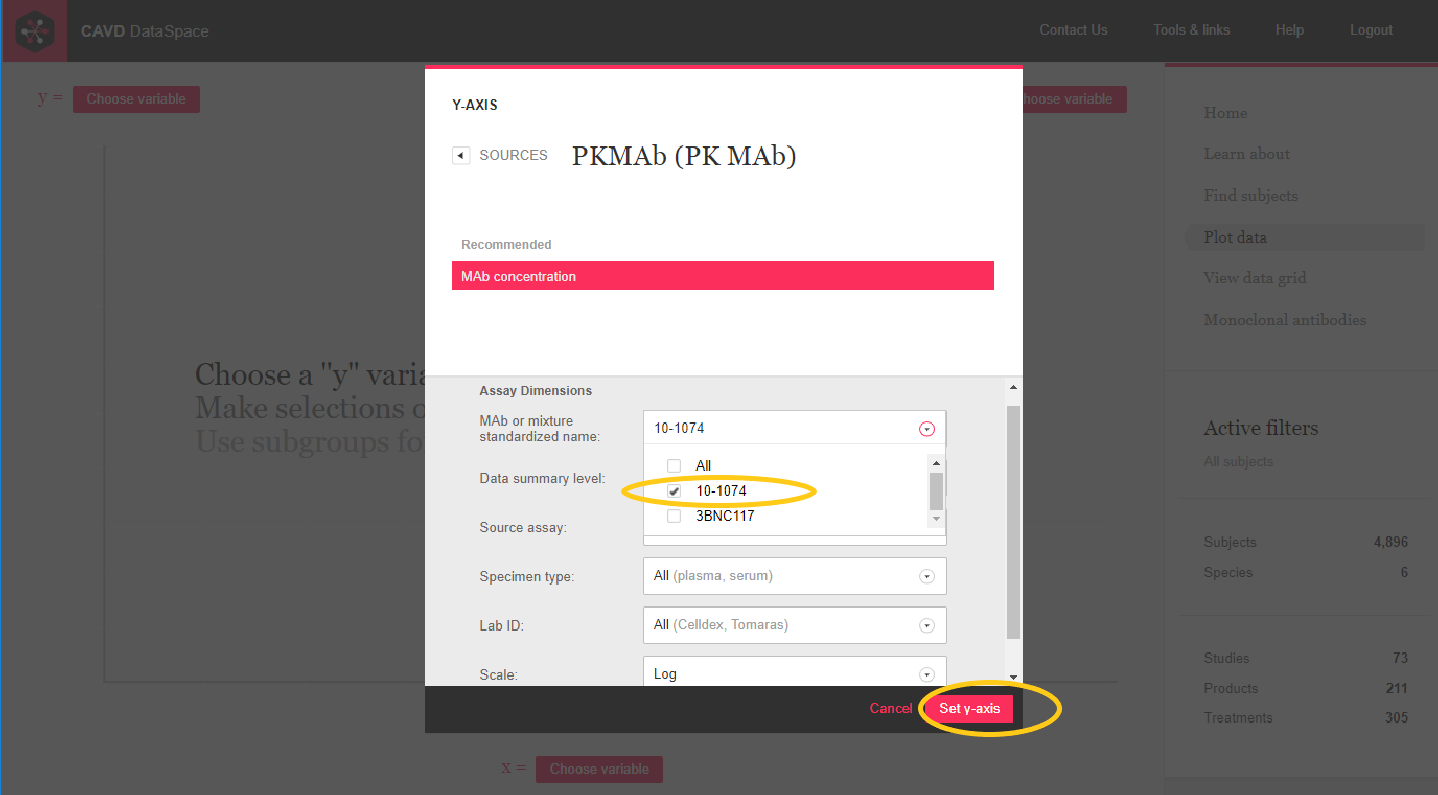
Step 3: Choose Study days from the Time points source to plot on the x-axis.
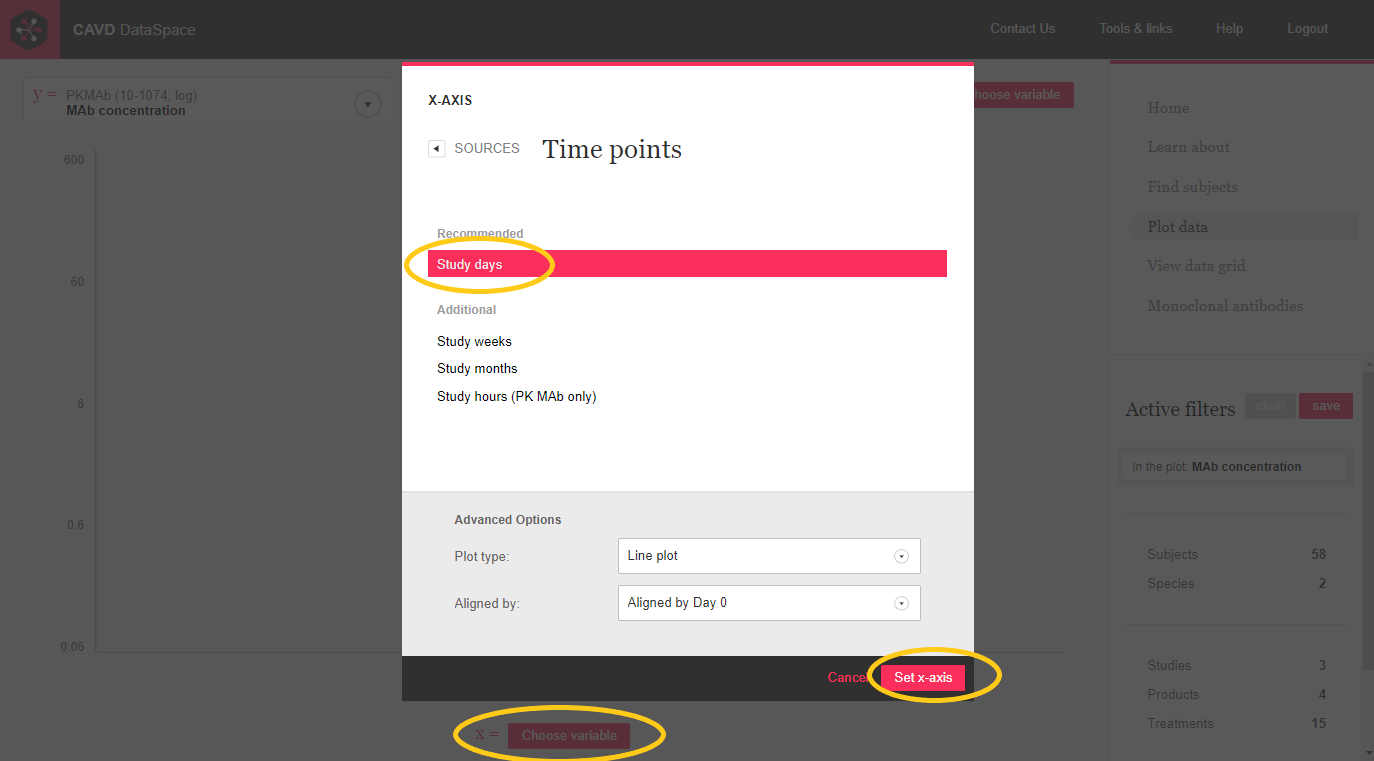
Step 4: Hmm.... more than one species and study. Let's filter to the human trial, CAVD 583.
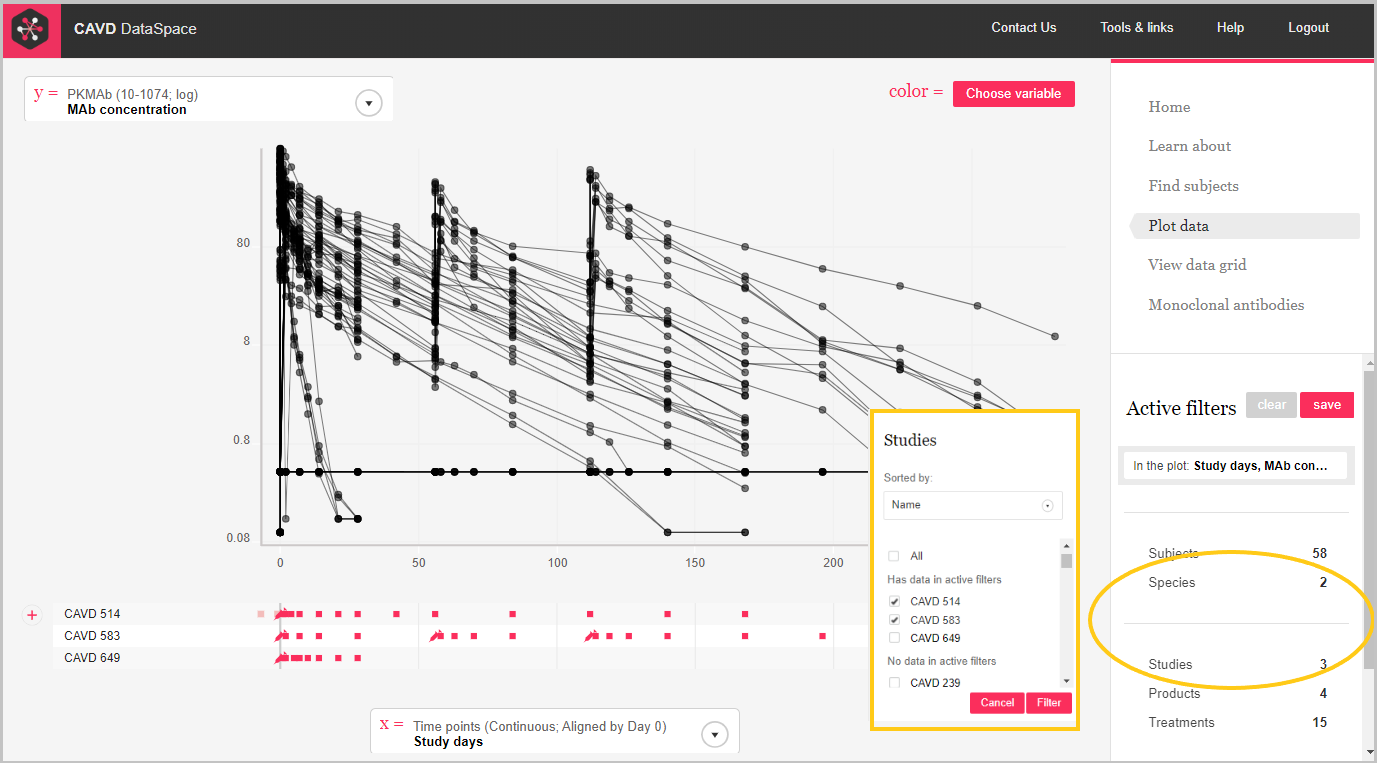
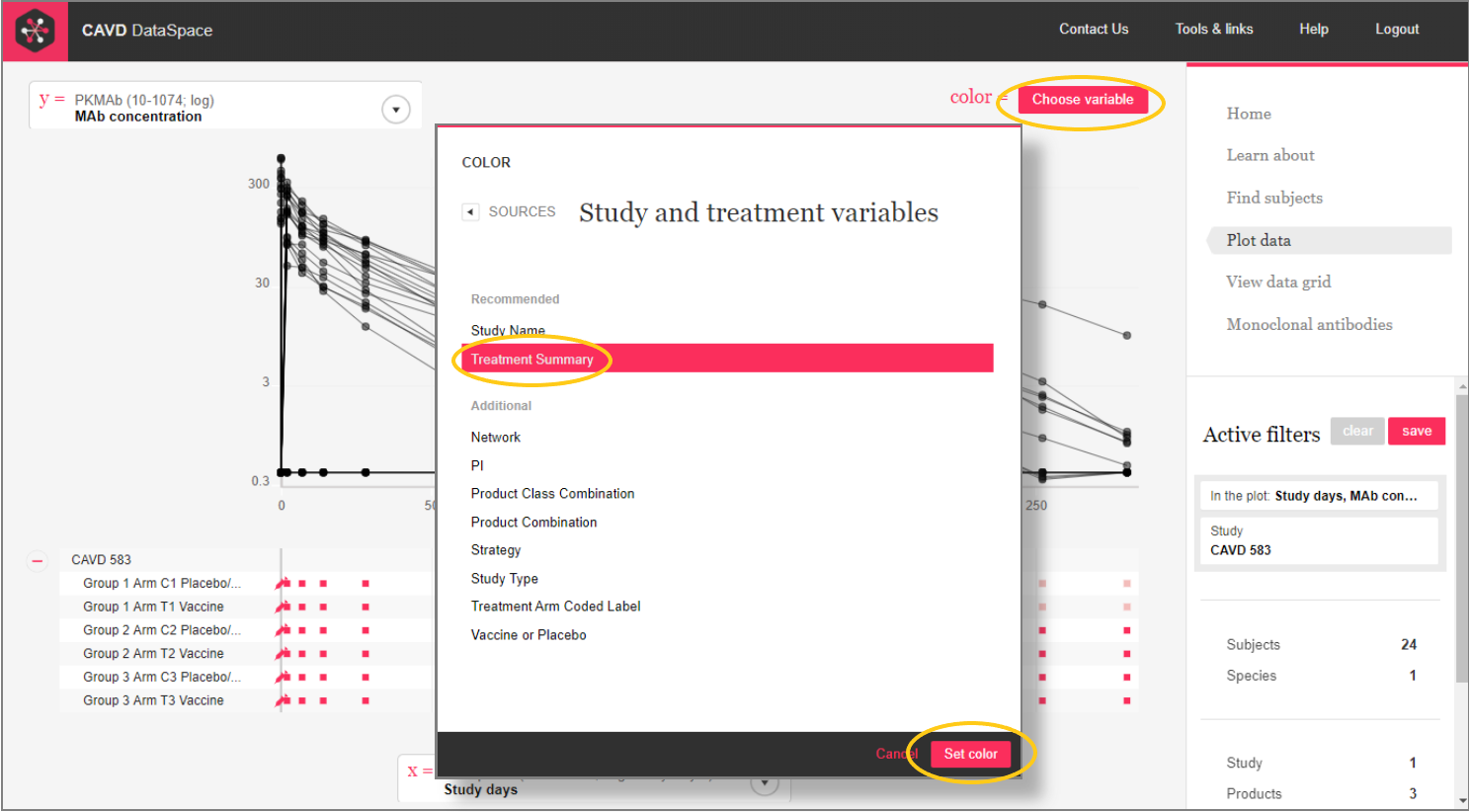
Step 5: We can use color as another dimension.
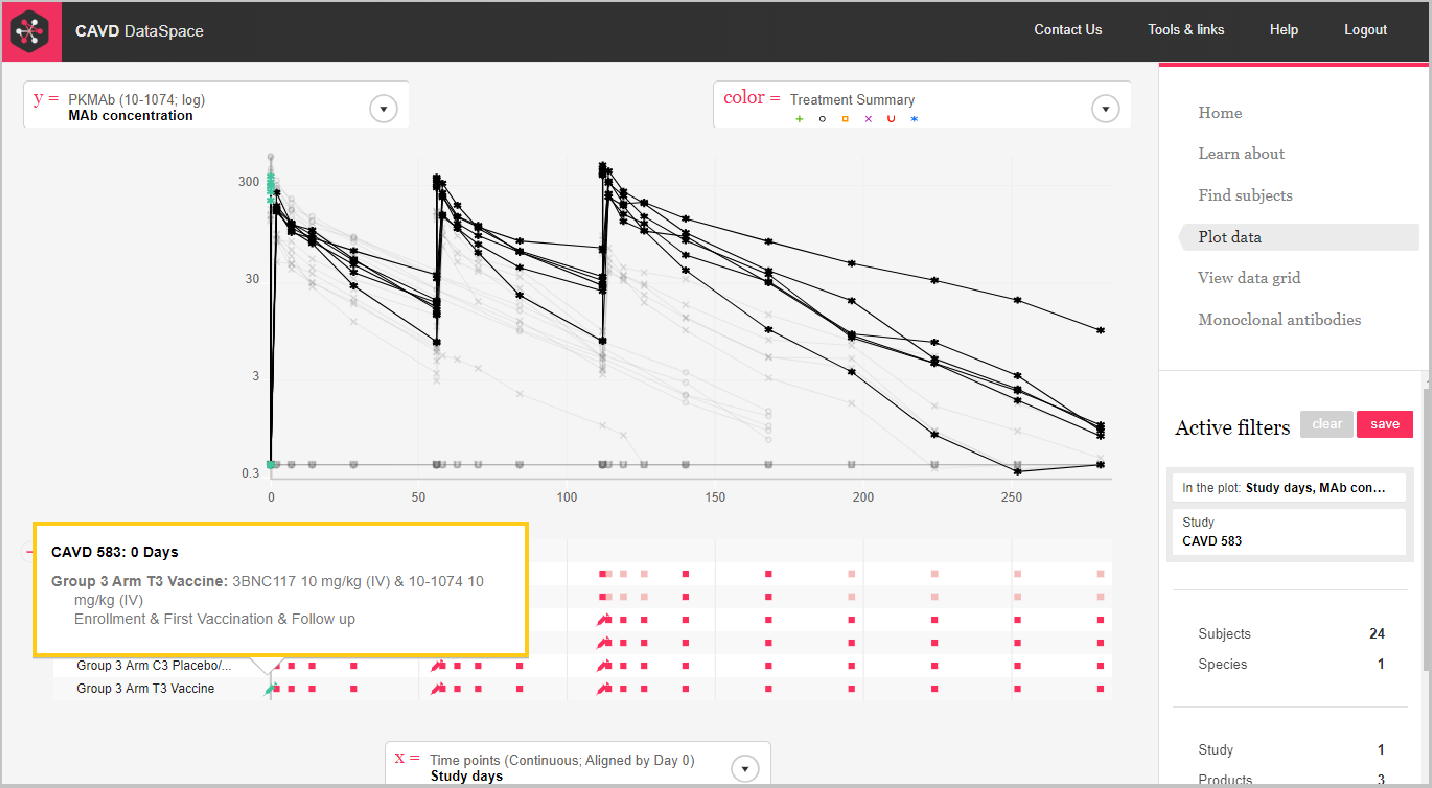
Step 5: Hover over the color box to see the key.
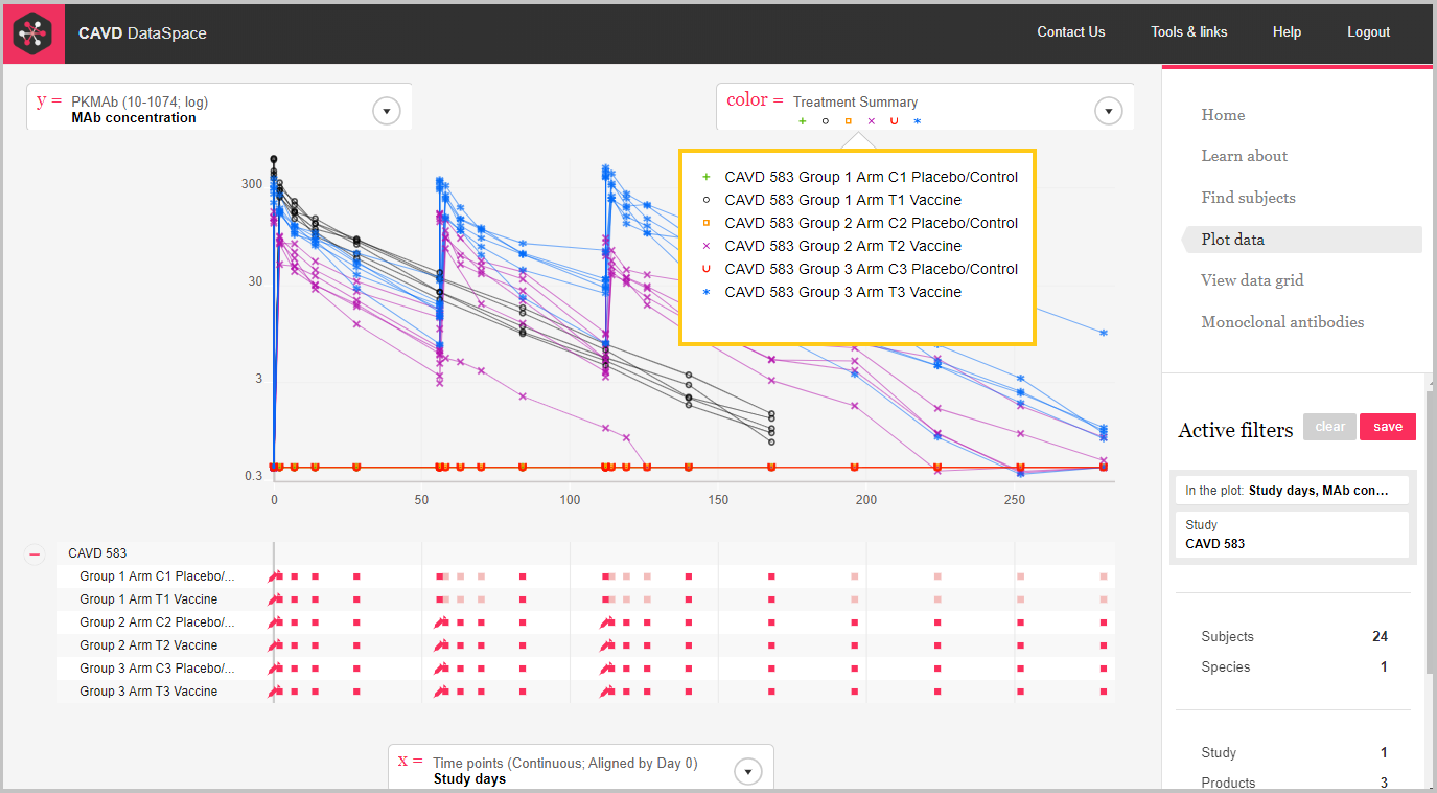
Remember you can hover over the syringe icon to see what products and doses were administered.
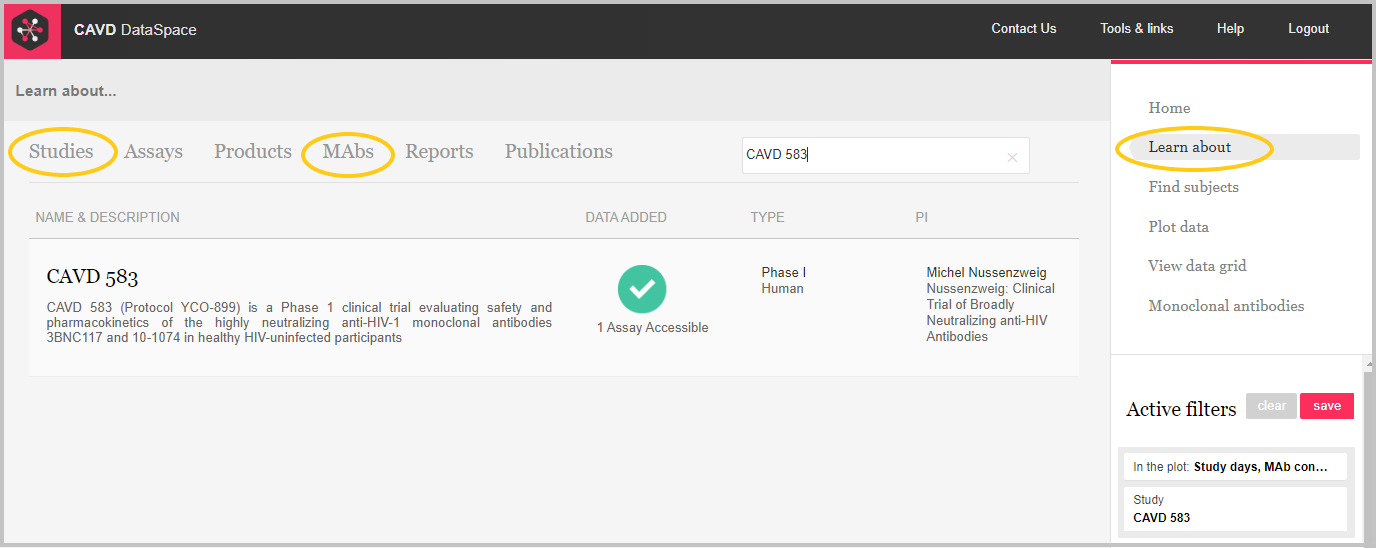
Go to Learn about to find more information about the CAVD 583 study or the 10-1074 and 3BNC117 mAbs.
What's next?
There are 13 CAVD and HVTN studies with PK data currently in the pipeline. Data will be loaded once the studies are complete.
We're continuing to refine and add to this new feature. Soon you'll be able to plot and compare concentrations for more than one mAb at a time. Adding the mAb name as color will help to highlight the differences.
We'd love to hear from you, so send us your feedback. Just click the Contact Us button at the top of the page or email us at [email protected].